CLEM-SIMS as a new method for resin-embedded samples
Abstract
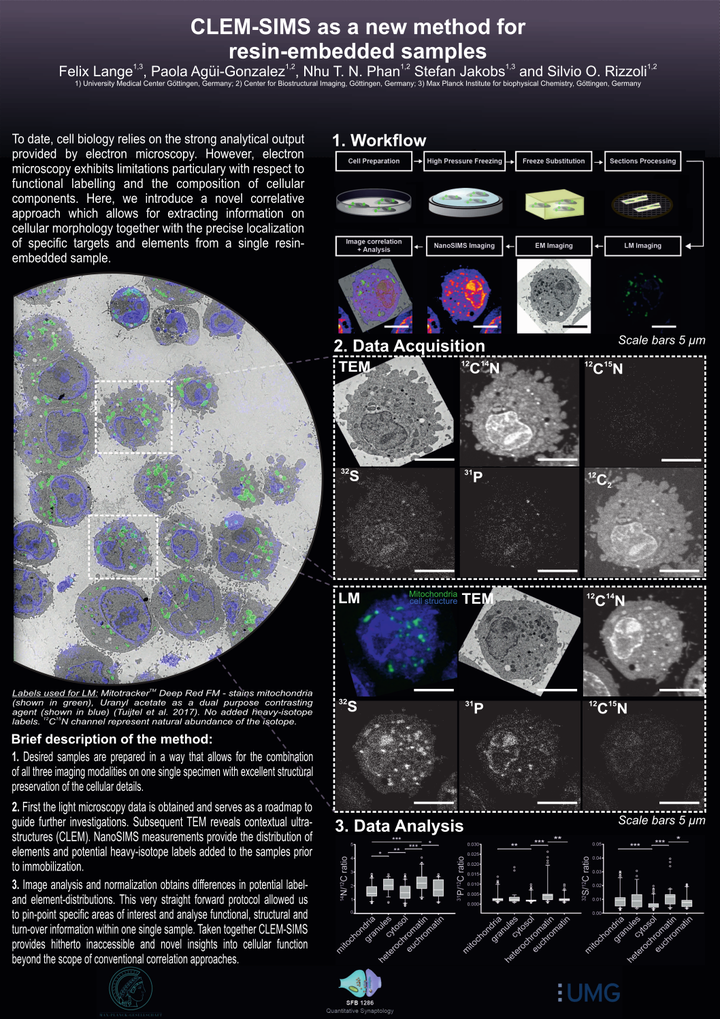
Cellular structure and function are currently investigated by a variety of imaging techniques, with resolutions ranging from sub-nanometer to millimetres. The best approaches to an understanding of the cellular structure are typically connected to the use of electron microscopy, in which dense cellular elements are visualised with high precision. A downside to this approach is that specific organelles are typically identified only based on their morphology, since most electron microscopy applications are performed without labelling the organelles in a specific fashion. A correlation with fluorescence microscopy (CLEM) enables the investigation of the location and function of specific cellular components by combining the information of both imaging modalities. While CLEM allows to re-assign functional information with decent precision within the corresponding ultrastructural context, it is still unable to address the chemical composition of cells, which is the domain of secondary ion mass spectrometry (SIMS). As SIMS is inherently unable to pinpoint specific cellular components, it has been correlated in several studies to either electron or fluorescence microscopy. However, only a correlation of all three techniques (CLEM-SIMS) would result in an optimal investigation of cell structure, function and composition. We generated here a protocol for CLEM-SIMS, based on transmission electron microscopy (TEM), conventional epifluorescence microscopy and nanoSIMS. The protocol was easily applied, and enabled the use of the three technologies at maximal performance parameters that can be carried out on a single prepared specimen. We suggest that CLEM-SIMS will provide substantial information that is currently beyond the possibilities of simpler correlative techniques.